A recent POP performance assessment investigated the performance of BayPass, a Fortran population genomics code parallelised with OpenMP. BayPass is able to identify genetic markers subjected to selection and associated with population-specific covariates.
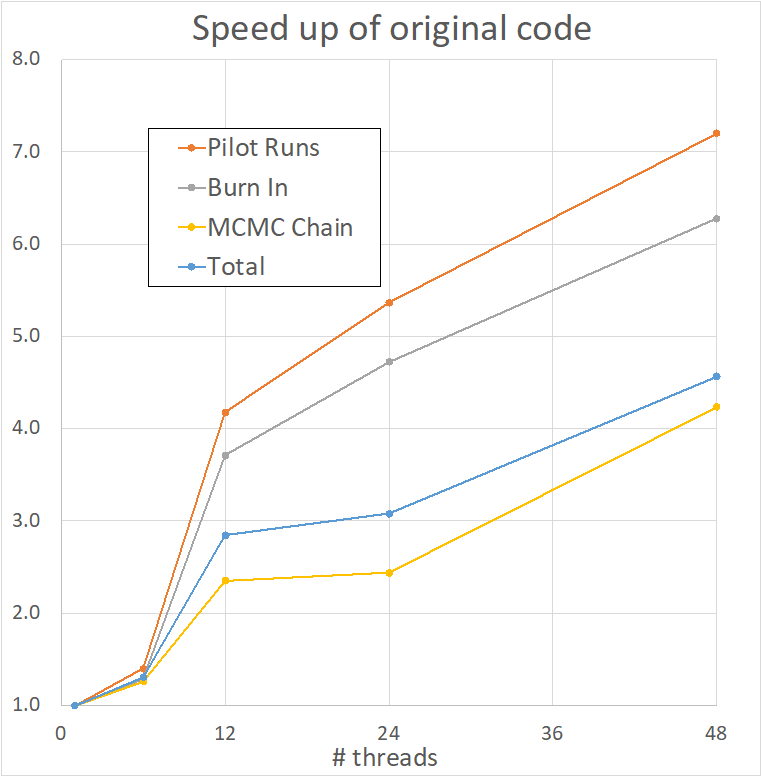 A detailed analysis of the different stages of the computation was undertaken by the POP team for both the Intel and GCC Fortran compilers. This revealed significant issues with IPC scaling, i.e. the number of useful instructions per cycle was reducing as the thread count increased, impacting parallel performance. The POP team undertook a thorough code review, in order to make specific recommendations on how to address this problem.
A detailed analysis of the different stages of the computation was undertaken by the POP team for both the Intel and GCC Fortran compilers. This revealed significant issues with IPC scaling, i.e. the number of useful instructions per cycle was reducing as the thread count increased, impacting parallel performance. The POP team undertook a thorough code review, in order to make specific recommendations on how to address this problem.
After implementing POP’s suggestions, the performance of BayPass was significantly improved. Dr Mathieu Gautier, the code developer said, "I am already able to see that the performance has been clearly and very highly improved. In some instances, the computation time is almost halved!"
-- Jonathan Boyle (NAG)
