A collaboration between POP and PerMedCoE started with the performance assessment of PhysiCell.
 PhysiCell is an open source, agent-based modeling framework for 3D multicellular simulations widely used in tissue and cancer biology to simulate the effect of genetic and environmental perturbations in cancer progression. It builds upon a multi-substrate diffusion-reaction solver to link cell phenotype to multiple substrates, such as nutrients and signaling factors. The application is written in C++ and is parallelized with OpenMP using parallel loops with a static schedule. Its computation cost scales linearly with the number of cells[1]. For the analysis, we used an experiment based on a heterogeneous 3D environment, which is used in PerMedCoE for performance studies and porting of PhysiCell.
PhysiCell is an open source, agent-based modeling framework for 3D multicellular simulations widely used in tissue and cancer biology to simulate the effect of genetic and environmental perturbations in cancer progression. It builds upon a multi-substrate diffusion-reaction solver to link cell phenotype to multiple substrates, such as nutrients and signaling factors. The application is written in C++ and is parallelized with OpenMP using parallel loops with a static schedule. Its computation cost scales linearly with the number of cells[1]. For the analysis, we used an experiment based on a heterogeneous 3D environment, which is used in PerMedCoE for performance studies and porting of PhysiCell.
“POP analysis has been tremendously useful to prioritize future developments and the scaling of PhysiCell and its derivative programs“ says Arnau Montagud, one of PhysiCell developers at the Life Science department at BSC.
To do our analysis, we focused on one iteration of the application, which performs 4 main different functions/tasks:
- Update the biochemical environment
- Update cell parameters, advance in the cell cycle, and change the cell’s volume
- Compute the new substrates’ gradient vectors
- Calculate the cell velocities vectors in accordance with its neighbors
Following the POP methodology and using the BSC tools (Extrae, Paraver, and BasicAnalysis suite) we studied the performance and scalability of the application, trying to identify the main factors limiting scalability. In our first execution of the application, we rapidly observed which parallel region has the major impact in the execution time. Figure 1 depicts a series of Paraver traces corresponding to executions from 2 to 48 threads, all at the same time scale. The big dark red region at the far right , which corresponds to the 4th task in the iteration as described before, takes most of the time and does not scale correctly.
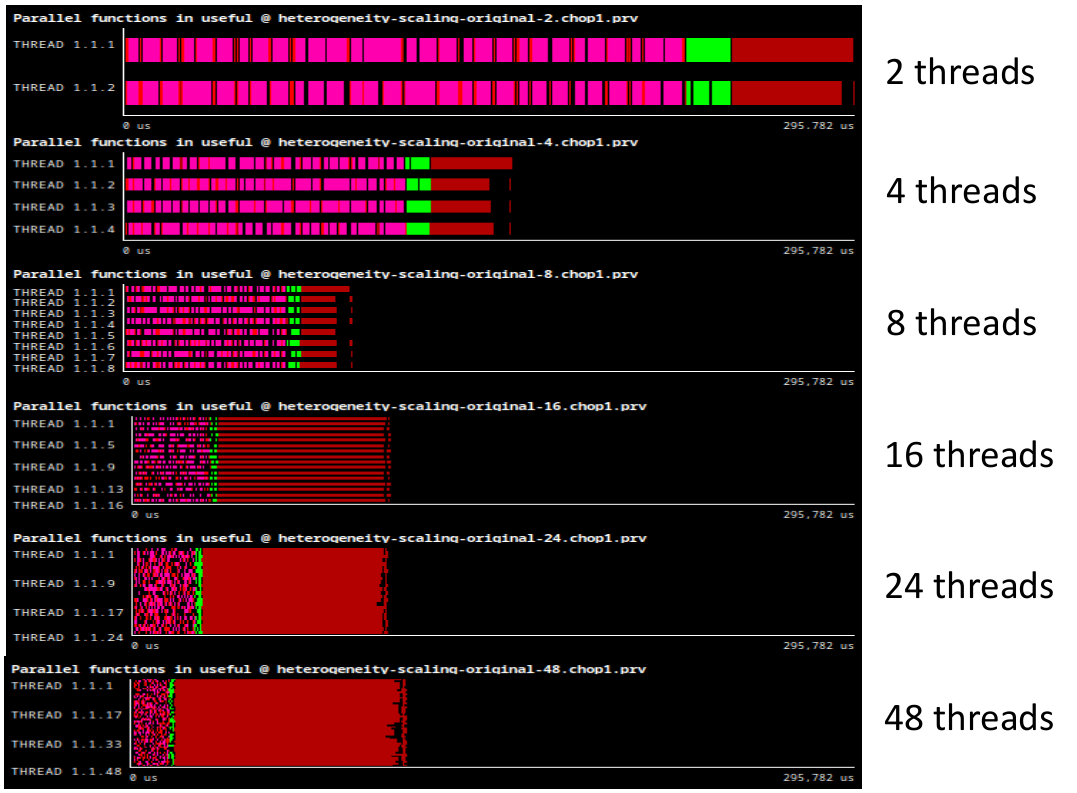
Figure 1: Execution trace of one iteration of PhysiCell with 2, 4, 8, 16, 24 and 48 OpenMP threads. Colors represent parallel functions. Y axis represent different threads. X axis represent time. All traces are depicted at the same time scale.
The detailed performance analysis led us to inspect a region of code where we discovered an unusually high density of memory allocations and deallocations due to the overloading of standard operators. Specifically, there are custom operators for the type vector<double> because the application usually operates with elements with 3 components, and vector addition element by element or vector by scalar multiplication are typical operations. The problem is the number of malloc and free calls that the runtime of C++ performs in this process. See Figure 2 and 3 as examples of multiple memory allocations and deallocations for a single operation.

Figure 2: Example of code line that does an overloaded addition and multiplication.

Figure 3: Implementation of the overloading of + operator for type vector<double>
This high numbers of memory allocations present a bottleneck when process scales to more than 8 threads, as the management of dynamic memory usually needs synchronization points. To solve this problem, we integrated a new library capable of handling memory allocations with concurrency in an efficient manner. Jemalloc is "a general purpose malloc(3) implementation that emphasizes fragmentation avoidance and scalable concurrency support"[2]. It can be integrated easily to any code by preloading the library, with LD_PRELOAD in Linux for example. After applying this solution to PhysiCell, we executed the same experiment and analyzed it with the same tools described before and found that we reduced the original run time by 31% for the case of 48 threads, or 27% reduction of the run time when using 24 threads. We also saw that the strong scalability is improved, going from a speedup of 25.4x (original malloc version) to 31.9x (Jemalloc version) for 48 threads (Figure 4).
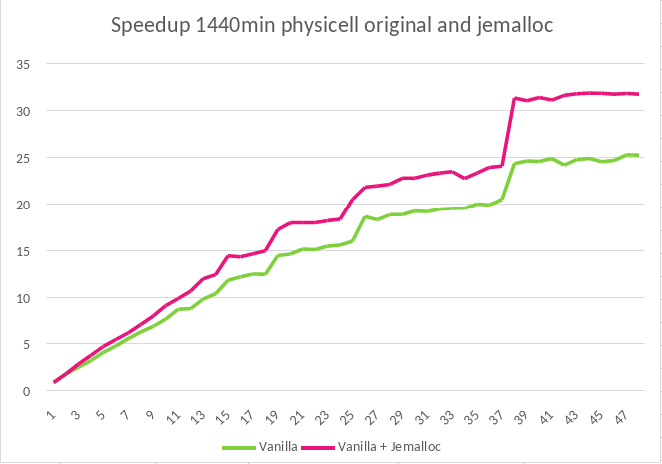
Figure 4: Speed-up comparison of PhysiCell running with two different memory allocation libraries, malloc and jemalloc.
[1] Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P (2018) PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput Biol 14(2): e1005991. https://doi.org/10.1371/journal.pcbi.1005991
[2] Jemalloc website. http://jemalloc.net
