HemeLB is an open-source lattice-Boltzmann code for simulation of large-scale three-dimensional fluid flow in complex sparse geometries such as those found in vascular networks. It is written in C++ using MPI by UCL and developed within the EU H2020 HPC CompBioMed Centre of Excellence for computational biomedicine as their flagship code.
Previous POP performance assessments of HemeLB with cerebral arterial circle of Willis geometry datasets on the Archer Cray XC30 up to 96,000 MPI processes (4,000 compute nodes) and Blue Waters Cray XE up to 239,615 MPI processes (18,432 compute nodes) confirmed the inherent scalability of the code.
To investigate the readiness of a more recent version of HemeLB for imminent exascale simulations, a new POP performance assessment was done with a higher-resolution 21.15 GiB testcase on the SuperMUC-NG Lenovo ThinkSystem SD650 at LRZ comprising 6,480 dual 24-core Intel Xeon Platinum 'Skylake' processor compute nodes. Although the code always ran correctly when sufficient memory was available, several compute nodes with notably inferior memory access performance were identified and subsequently explicitly avoided.
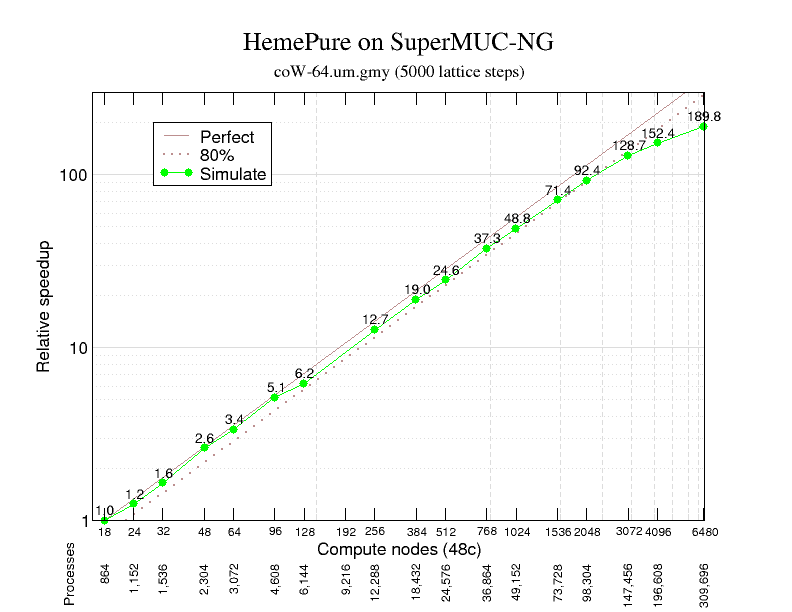
Strong scaling was examined with up to 309,696 MPI processes (on 6,452 compute nodes) using the Scalasca toolset. Compared to the smallest configuration that could be run using 864 processes (on 18 large-memory compute nodes each with 768 GiB), 190x speed-up was delivered with 80% scaling efficiency maintained to over 100,000 processes for the simulation phase.
Non-blocking MPI point-to-point message-passing ensures excellent communication efficiency, whereas load balance is somewhat variable but remains good at all scales. Computation scaling is excellent to over 50,000 processes before progressively deteriorating due to extra instruction overhead from local grid block surface to volume issues which are only partially compensated by improving IPC.
The summary of the performance assessment has further details. Ongoing work in collaboration with the E-CAM CoE is evaluating a load-balancing library (ALL) that they have developed, and more generally in preparation of coupled simulations using patient-specific geometries of the entire arterial and venous trees.
